4D intravital imaging at single cell resolution
Methods to analyze 4D single cell dynamics in a living mouse.
In order to study the dynamics of cell growth and cell cycle progression in an adult animal, I developed an experimental system to directly observe and track hundreds of single cells in a living mouse, over long periods of time, in 4D. (Xie et al., 2024)
Most of the functionality described in this page is available at this repository.
Intravital imaging of mouse skin
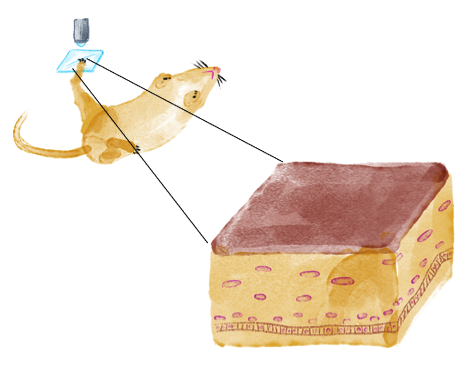
The methods to image mouse cells were pioneered by Valentina Greco’s group, who established protocols to perform the longitudinal re-imaging of the same tissue regions, and has made available many reporter mouse lines generated by her group.
This powerful technique uses two-photon microscopy, which can penetrate deeper into tissues compared to single-photon excitation. I can directly image fluorescent mouse lines that express reporters for cellular structures (e.g. nucleus, cell boundary) and cell states (e.g. cell cycle phase, cell type), and produces three-dimensional images of a small voxel of the living epidermis.
I then use various landmarking techniques to navigate back to the same exact tissue region in the same mouse over many imaging sessions, allowing the mouse to return to its normal housing and activity in between short imaging sessions.
This technique can collect time-lapsed ‘3D snapshots’ of the same piece of skin tissue over many days, if not months.
Image processing: 3D registration
Although we have these 3D snapshots of the same region, they are not pixel-perfect. There are always slight variations in position and angle from timepoint to timepoint. Therefore, I built tools to register timepoints in 3D, and generate a final ‘aligned movie’ with fixed coordinates. I am currently working on publishing a napari plugin of this functionality.
Single cell analysis: 3D segmentation
Segmentation in 3D remains challenging to do accurately and at scale. I generated training data and trained deep learning models (based on stardist3D, plantseg, and cellpose working in 3D stitching mode) to perform 3D boundary segmentation with high fidelity. These models can be deployed for analyzing new images I generate, allowing me to process datasets much faster.
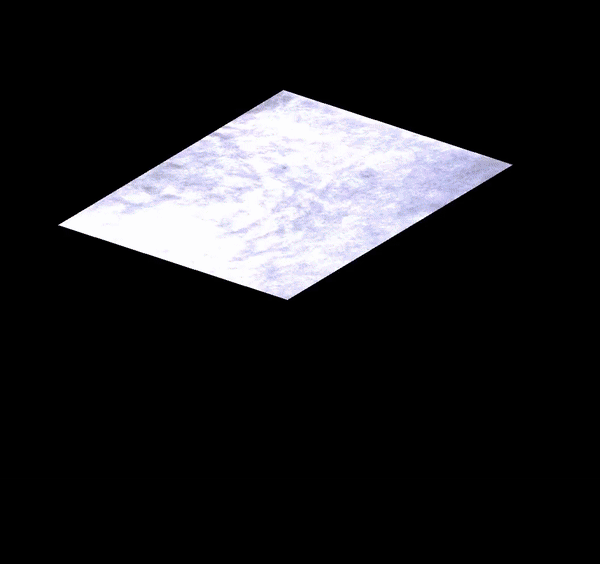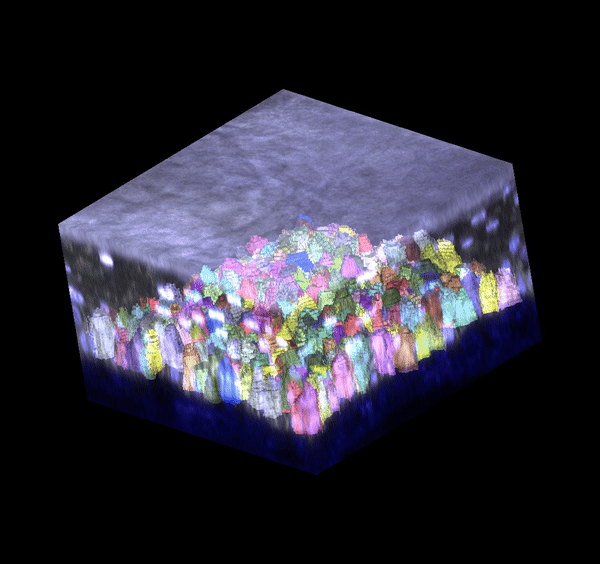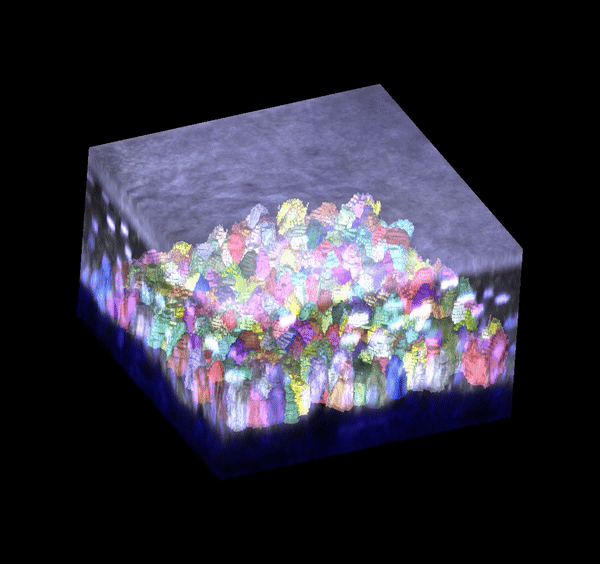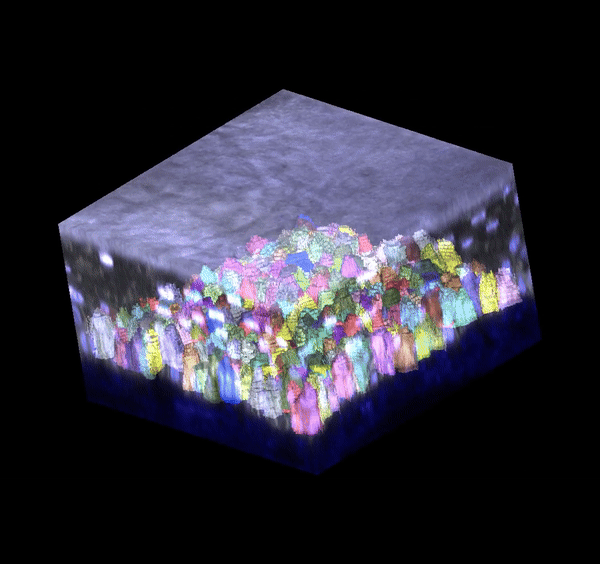
Single cell analysis: 3D tracking
Cell tracking poses an even larger challenge compared to segmentation, especially in 3D. Currently, I am performing semi-automated cell tracking in 3D using the Mastodon lineage visualization and tracking, and I have developed software to interface with Mastodon outputs and integrate them into the rest of the analysis pipeline.
This produces high-fidelity tracking of skin stem cells in my movies.
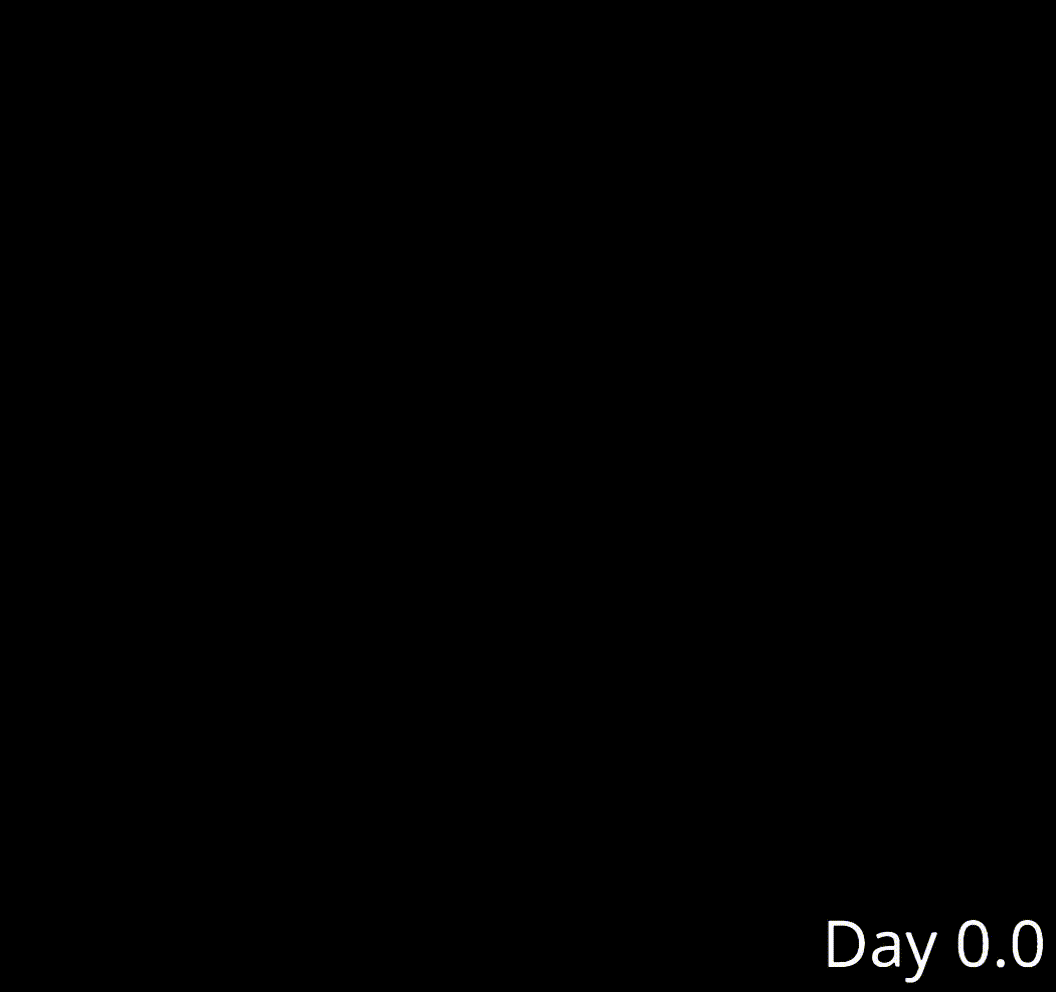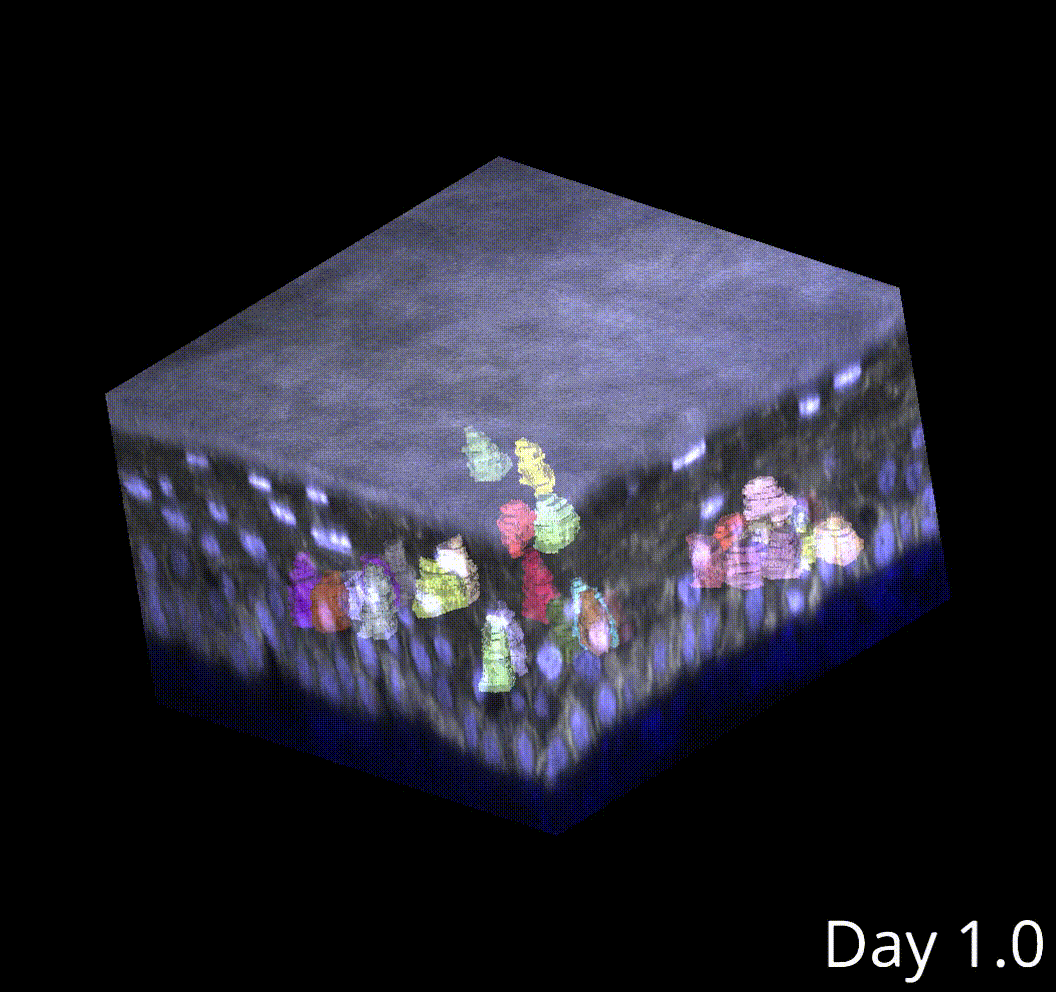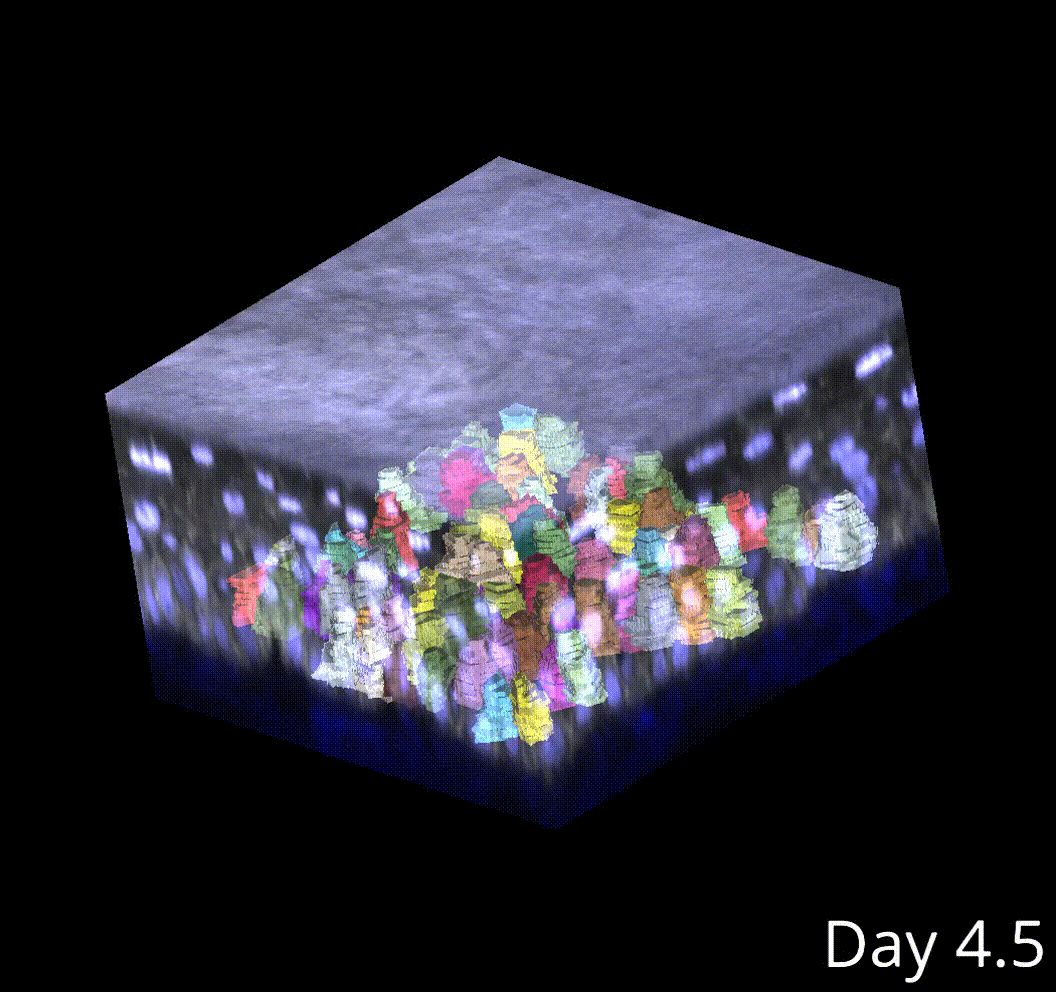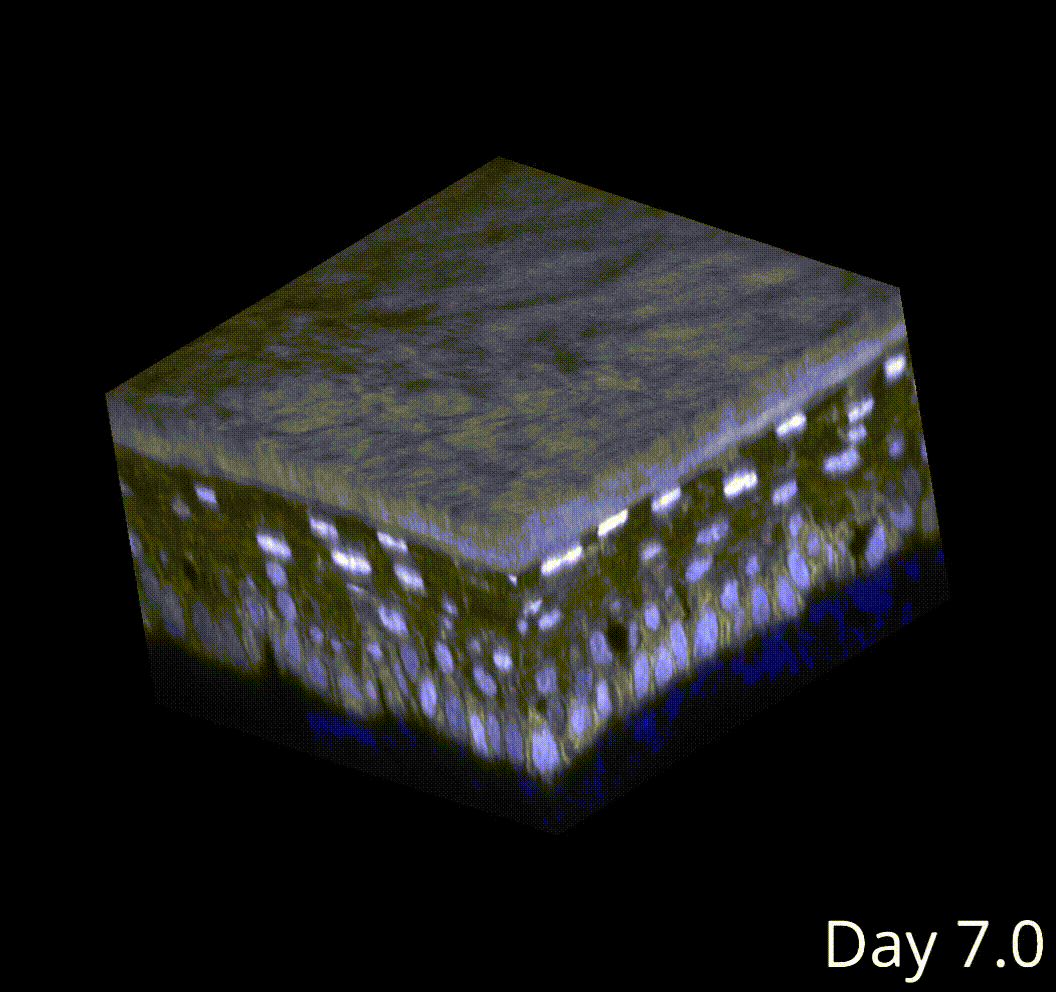
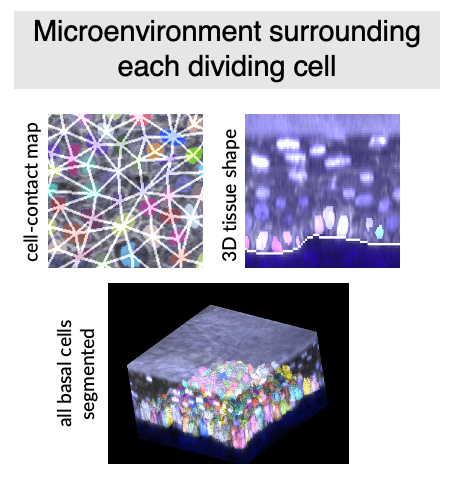
3D microenvironment analysis
In order to analyze how the microenvironment around each cell changes over time, I developed a computational framework to:
- encode cell-cell contact maps in 3D: by automatically inferring what cells are touching from 3D segmentations and defining a graph-based encoding of each cell’s “neighborhood”
- extract tissue-level geometry and its changes: by fitting 3D meshes of the epidermis-dermis interface and performing geometric operations on these meshes
- extract features of the extracellular collagen matrix that is in contact with each cell: by analyzing the anisotropies of the second harmonic generation signal generated by collagen in the dermis
Combining this framework with high-fidelity 3D single cell segmentation and tracking yields a rich dataset that has unprecedented insight into the cellular dynamics that underly how an in vivo tissue evolves over time.